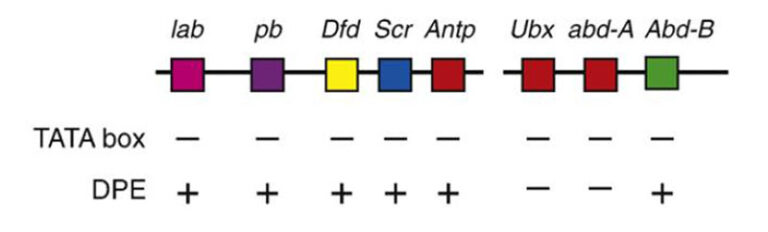
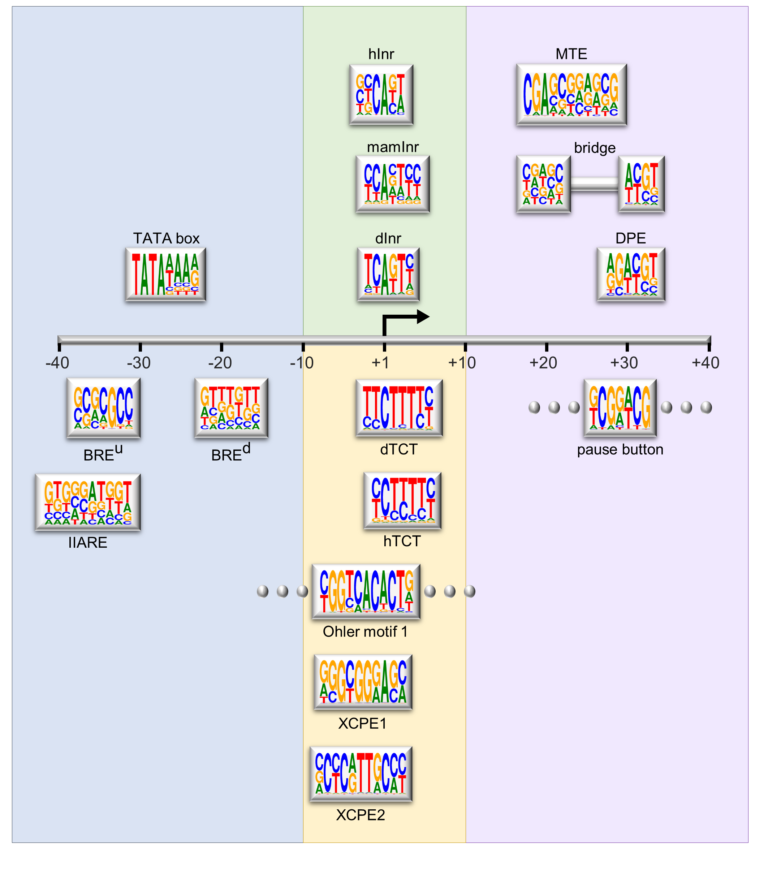
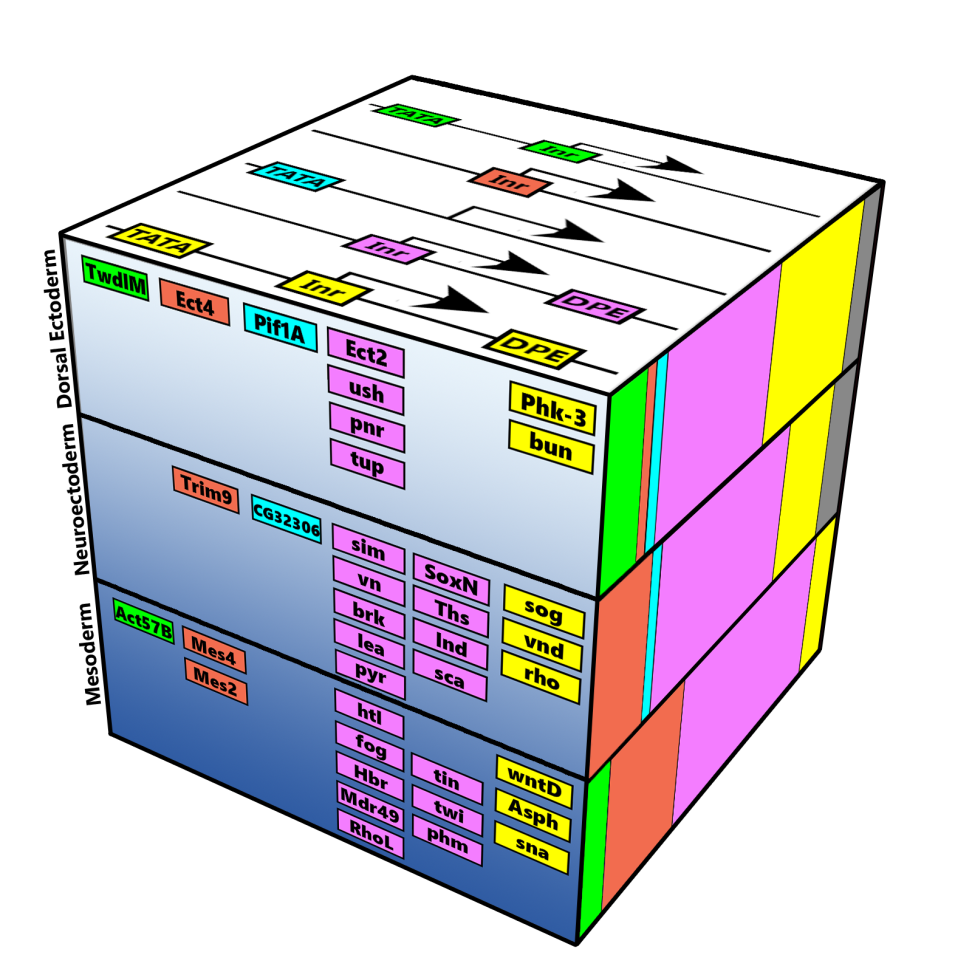
Schematic representation of the major core promoter elements.
Full information is available here
To achieve these goals, we integrate techniques from molecular biology, biochemistry, cell biology, bioinformatics, and developmental biology.
We employ Drosophila melanogaster (the fruit fly) as our model organism due to its long-standing contribution/use in genetics and developmental biology, as well as its molecular similarities to humans.
Remarkably, many regulatory mechanisms in flies have parallels in human biology, enabling insights that extend beyond Drosophila. By examining how transcriptional regulation guides development in the fly, we gain valuable knowledge relevant to human biology.
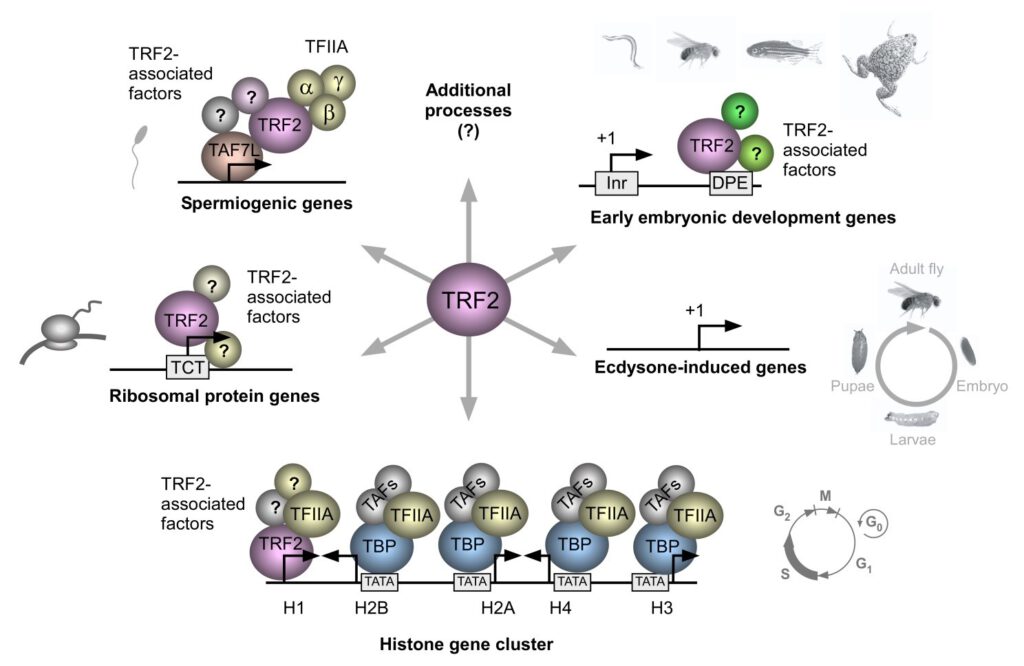
Our studies focus on the molecular mechanisms underlying the formation of diverse body structures during development and emphasize the broader implications of core promoter diversity. This work is critical not only for understanding development but also for illuminating processes linked to diseases such as cancer, where transcriptional regulation can become dysregulated.
Together, our analyses of core promoters and the proteins that bind and regulate them are essential for deciphering fundamental molecular and mechanistic aspects of transcription. These insights contribute to the broader understanding of fundamental biological processes, including development and differentiation.